Regulatory analysis of the C. elegans genome with spatiotemporal resolution
2014y
Science
Nature
Carlos Araya
Araya C.L., et al. Nature 512, 400–405 (2014)
Abstract
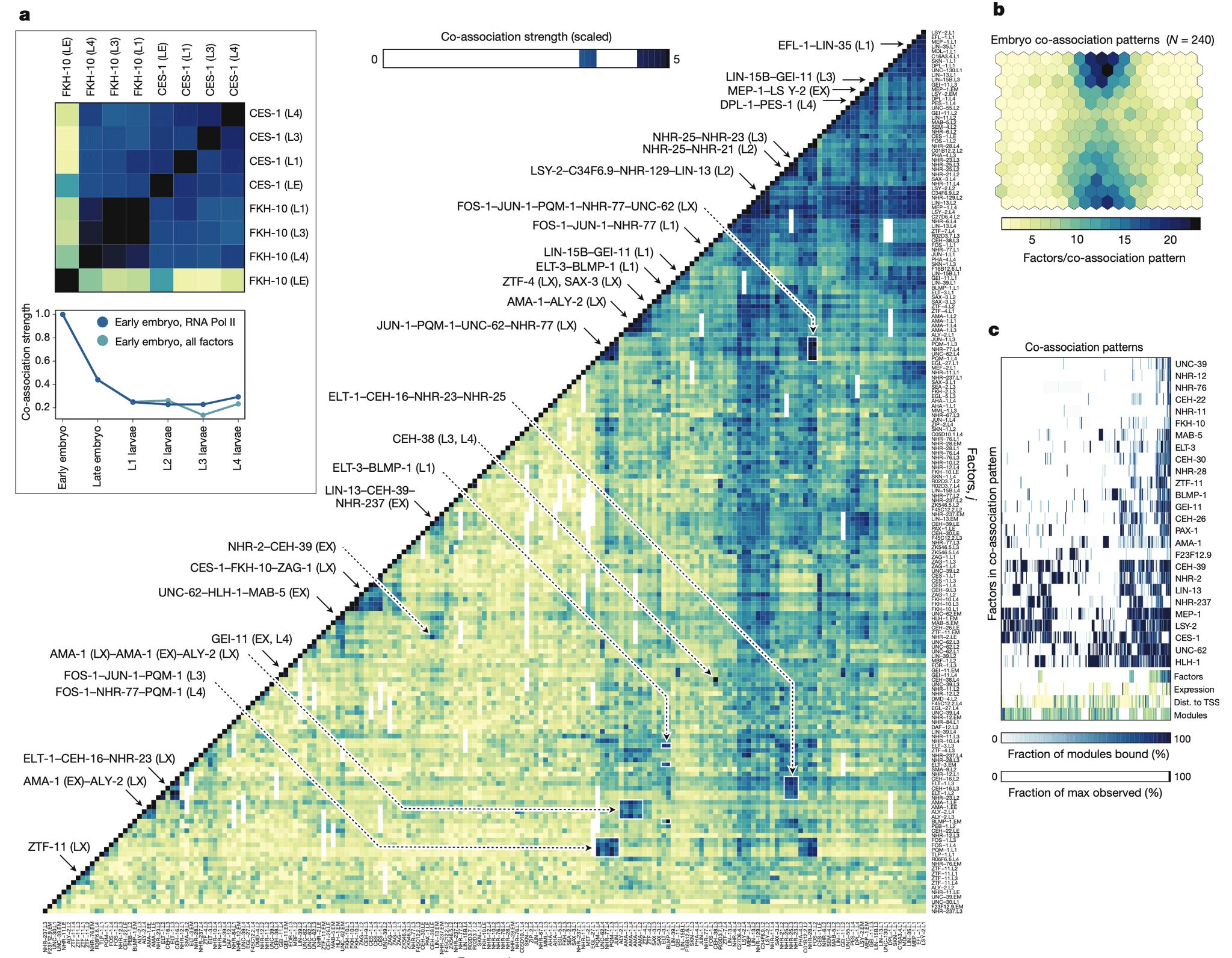
Discovering the structure and dynamics of transcriptional regulatory events in the genome with cellular and temporal resolution is crucial to understanding the regulatory underpinnings of development and disease. We determined the genomic distribution of binding sites for 92 transcription factors and regulatory proteins across multiple stages of Caenorhabditis elegans development by performing 241 ChIP-seq (chromatin immunoprecipitation followed by sequencing) experiments. Integration of regulatory binding and cellular-resolution expression data produced a spatiotemporally resolved metazoan transcription factor binding map. Using this map, we explore developmental regulatory circuits that encode combinatorial logic at the levels of co-binding and co-expression of transcription factors, characterizing the genomic coverage and clustering of regulatory binding, the binding preferences of, and biological processes regulated by, transcription factors, the global transcription factor co-associations and genomic subdomains that suggest shared patterns of regulation, and identifying key transcription factors and transcription factor co-associations for fate specification of individual lineages and cell types.
Experiences
Find frequently asked questions about my products:
More Projects
Questions?
Reach out and I'll get back to you in a few hours.