A fundamental protein property, thermodynamic stability, revealed solely from large-scale protein fitness data
2012y
Science
PNAS
Carlos Araya, Douglas Fowler
Araya C.L.*, Fowler D.M.*, et al. Proceedings of the National Academy of Science 109 (42) 16858-16863 (2012)
Abstract
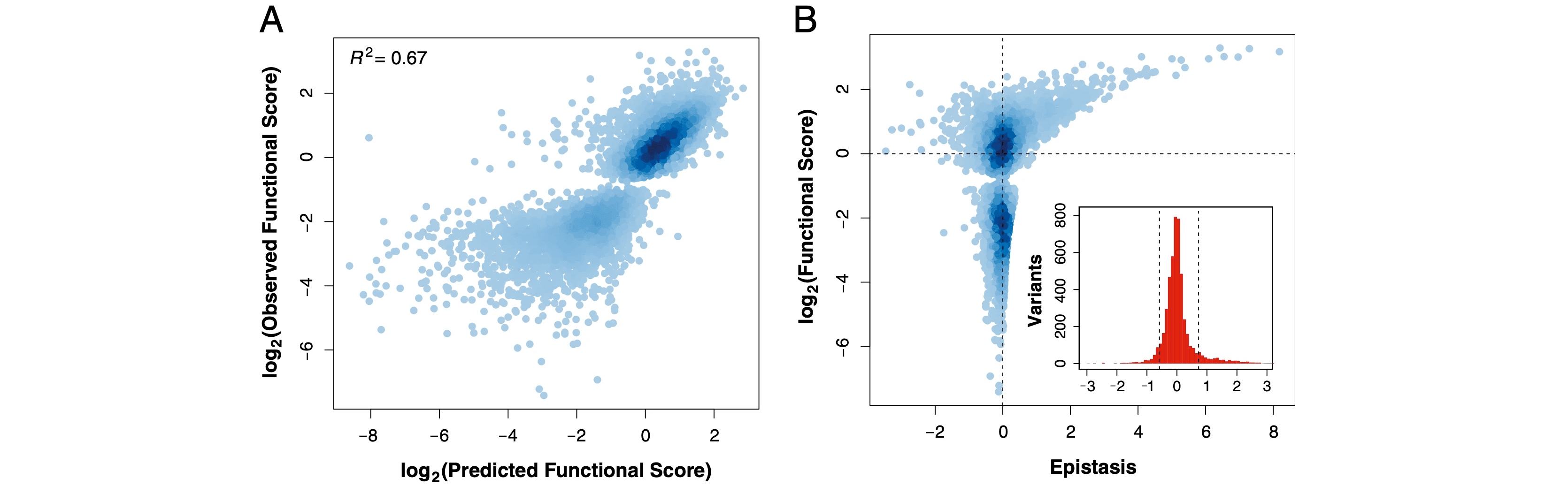
The ability of a protein to carry out a given function results from fundamental physicochemical properties that include the protein’s structure, mechanism of action and thermostability. Traditional approaches to study these protein properties have typically required the direct measurement of the property of interest, oftentimes a laborious undertaking. Recent technological developments have enabled the rapid and large-scale quantification of the relationship between the sequence of a protein and its fitness in a simple assay, such as binding. Here, we measure the fitness of 47,000 variants of a WW domain to bind to a peptide ligand and use these measurements to identify stabilizing mutations without directly assaying stability. Our approach is rooted in the well-established concept that stabilizing mutations can permit the acquisition of other, destabilizing mutations that improve function. This phenomenon can be observed as epistasis, wherein multiple mutations combine with unpredictable fitness effects. We introduce an epistasis-based metric, “partner potentiation,” which identified 15 candidate stabilizing mutations. We tested six novel candidates by thermal denaturation and found two highly stabilizing mutations, one more stabilizing than any previously known mutation. Thus, physicochemical properties such as stability are latent within large-scale protein fitness data and can be revealed by systematic analysis. This approach represents a new direction in protein science, the use of large protein fitness datasets to uncover fundamental protein properties.
Experiences
Find frequently asked questions about my products:
More Projects
Questions?
Reach out and I'll get back to you in a few hours.